To our knowledge, we are unique among the Shock Centers in providing services that take advantage of the powerful model organisms C. elegans and S. cerevisiae. The center has a deep history of effectively using these organisms to investigate the fundamental biology of aging, and for testing hypotheses generated from humans and other study systems. We also continue to develop and use new technology with these organisms.
S. cerevisiae Chemostat Assays. Dr. Dunham continues to develop the yeast chemostat devices for aging studies. These devices grow large populations of aged yeast cells by magnetically binding a young population to the side of the device while daughter buds flow out of the chamber, enriching for older cells over time. Chemostat assays typically cost about $2000 per experiment, and can be performed with custom strains and media. These devices have been used to enrich for older cells for aging studies by grantees like Dr. Alaattin Kaya and Dr. Daniel Pollard, as well as internally for collaborations with the Protein Phenotypes of Aging Core.
S. cerevisiae dissection microscope loan program. For labs who would like to perform microdissection-based replicative lifespan assays, the core offers microscope loans and training for no cost.
C. elegans Lifespan Assays. Dr. Mendenhall leads the C. elegans services portion of the Core. Lifespan Assays are $1000 per assay, which includes over 100 control animals and over 100 experimental animals, and data analysis. Lifespan assays are typically performed in a semiautomated fashion on WormBots with FUDR added (WormBots robotically image animals in 12-well solid NGM plates), but can be assayed manually without the addition of FUDR if awardees would like to avoid the use of FUDR, as some lifespan effects depend on FUDR, which is only used in humans for cancer treatment, as it inhibits DNA synthesis. We have performed lifespan assays for several labs, such as Dr. Nick Lehbach.
C. elegans Genome Editing/Reporter Gene Construction. Precision Genome edits are typically $5000 per edit, with three independent edits typically produced and delivered to the awardee. Extrachromosomal arrays are often expressed at higher levels and cost less, depending on the nature of the construct. There are advantages and drawbacks to each approach, and the optimal approach choice depends on the goals of the study. We have made reporter genes for several prominent worm aging labs including the Driscoll Lab and Leiser Lab.
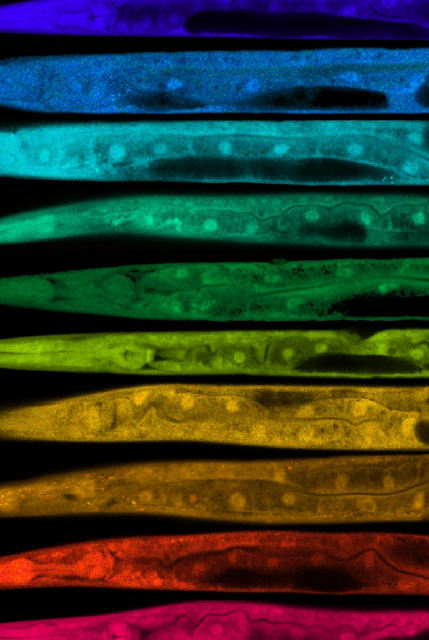
Quantitative Light Microscopy. Our core has unique expertise in cell biology, C. elegans anatomy and physiology, and quantitative light imaging methods Dr. Mendenhall developed. Quantitative Fluorescent Confocal Light Microscopy Assays are typically $1-2000 per assay, which includes 5-10 control animals and 5-10 experimental animals, quantitatively imaged at cellular or subcellular resolution, depending on the design of the study. For yeast or cultured mammalian cells, we will typically image dozens to hundreds of cells per group, often in a few different fields of view or from different wells. We have performed imaging assays for labs such as the McCormick Lab and the Leiser Lab.
Allele Resolution Sequencing: Silencing and Expression with Age. We have developed a new technical capability to study changes in the genome with age, especially given our recent findings on widespread allele expression bias and huge changes in silencing in the brain (Mariner et al 2025). In a recent partnership with Shock Center Grantee Dr. Alexander Gimelbrant and local second and third generation sequencing experts, Dr. Scott Kennedy and Dr. Danny Miller, we have set up a sequencing pipeline that generates haplotype resolution RNA expression and DNA sequence data, including accurate, nucleotide-resolution 5mC marks. Scientists can observe changes in inferred silencing with long read sequencing revealing differential methylation of promoters, and see that silencing at the RNA level in individual alleles with bulk tissue samples. The pipeline is currently up and running and complements our genetic and microscopic studies of aging. Data is rich but costs are relatively high. Human genome samples cost $3000 for long read sequencing at allele resolution and $1000 for RNA-sequencing at haplotype resolution, which requires the long read sequencing for measuring the SNPs in the sample. Costs may be reduced for targeted sequencing experiments focusing on 10-100 targets.
Core Personnel: The Core is led by Maitreya Dunham and Alex Mendenhall